Pandas Dataframes¶
This notebook demonstrates how systematic analysis of tally scores is possible using Pandas dataframes. A dataframe can be automatically generated using the Tally.get_pandas_dataframe(...) method. Furthermore, by linking the tally data in a statepoint file with geometry and material information from a summary file, the dataframe can be shown with user-supplied labels.
[1]:
import glob
from IPython.display import Image
import matplotlib.pyplot as plt
import scipy.stats
import numpy as np
import pandas as pd
import openmc
%matplotlib inline
Generate Input Files¶
First we need to define materials that will be used in the problem. We will create three materials for the fuel, water, and cladding of the fuel pin.
[2]:
# 1.6 enriched fuel
fuel = openmc.Material(name='1.6% Fuel')
fuel.set_density('g/cm3', 10.31341)
fuel.add_nuclide('U235', 3.7503e-4)
fuel.add_nuclide('U238', 2.2625e-2)
fuel.add_nuclide('O16', 4.6007e-2)
# borated water
water = openmc.Material(name='Borated Water')
water.set_density('g/cm3', 0.740582)
water.add_nuclide('H1', 4.9457e-2)
water.add_nuclide('O16', 2.4732e-2)
water.add_nuclide('B10', 8.0042e-6)
# zircaloy
zircaloy = openmc.Material(name='Zircaloy')
zircaloy.set_density('g/cm3', 6.55)
zircaloy.add_nuclide('Zr90', 7.2758e-3)
With our three materials, we can now create a materials file object that can be exported to an actual XML file.
[3]:
# Instantiate a Materials collection
materials = openmc.Materials([fuel, water, zircaloy])
# Export to "materials.xml"
materials.export_to_xml()
Now let’s move on to the geometry. This problem will be a square array of fuel pins for which we can use OpenMC’s lattice/universe feature. The basic universe will have three regions for the fuel, the clad, and the surrounding coolant. The first step is to create the bounding surfaces for fuel and clad, as well as the outer bounding surfaces of the problem.
[4]:
# Create cylinders for the fuel and clad
fuel_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, r=0.39218)
clad_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, r=0.45720)
# Create boundary planes to surround the geometry
# Use both reflective and vacuum boundaries to make life interesting
min_x = openmc.XPlane(x0=-10.71, boundary_type='reflective')
max_x = openmc.XPlane(x0=+10.71, boundary_type='vacuum')
min_y = openmc.YPlane(y0=-10.71, boundary_type='vacuum')
max_y = openmc.YPlane(y0=+10.71, boundary_type='reflective')
min_z = openmc.ZPlane(z0=-10.71, boundary_type='reflective')
max_z = openmc.ZPlane(z0=+10.71, boundary_type='reflective')
With the surfaces defined, we can now construct a fuel pin cell from cells that are defined by intersections of half-spaces created by the surfaces.
[5]:
# Create fuel Cell
fuel_cell = openmc.Cell(name='1.6% Fuel', fill=fuel,
region=-fuel_outer_radius)
# Create a clad Cell
clad_cell = openmc.Cell(name='1.6% Clad', fill=zircaloy)
clad_cell.region = +fuel_outer_radius & -clad_outer_radius
# Create a moderator Cell
moderator_cell = openmc.Cell(name='1.6% Moderator', fill=water,
region=+clad_outer_radius)
# Create a Universe to encapsulate a fuel pin
pin_cell_universe = openmc.Universe(name='1.6% Fuel Pin', cells=[
fuel_cell, clad_cell, moderator_cell
])
Using the pin cell universe, we can construct a 17x17 rectangular lattice with a 1.26 cm pitch.
[6]:
# Create fuel assembly Lattice
assembly = openmc.RectLattice(name='1.6% Fuel - 0BA')
assembly.pitch = (1.26, 1.26)
assembly.lower_left = [-1.26 * 17. / 2.0] * 2
assembly.universes = [[pin_cell_universe] * 17] * 17
OpenMC requires that there is a “root” universe. Let us create a root cell that is filled by the pin cell universe and then assign it to the root universe.
[7]:
# Create root Cell
root_cell = openmc.Cell(name='root cell', fill=assembly)
# Add boundary planes
root_cell.region = +min_x & -max_x & +min_y & -max_y & +min_z & -max_z
# Create root Universe
root_universe = openmc.Universe(name='root universe')
root_universe.add_cell(root_cell)
We now must create a geometry that is assigned a root universe and export it to XML.
[8]:
# Create Geometry and export to "geometry.xml"
geometry = openmc.Geometry(root_universe)
geometry.export_to_xml()
With the geometry and materials finished, we now just need to define simulation parameters. In this case, we will use 5 inactive batches and 15 minimum active batches each with 2500 particles. We also tell OpenMC to turn tally triggers on, which means it will keep running until some criterion on the uncertainty of tallies is reached.
[9]:
# OpenMC simulation parameters
min_batches = 20
max_batches = 200
inactive = 5
particles = 2500
# Instantiate a Settings object
settings = openmc.Settings()
settings.batches = min_batches
settings.inactive = inactive
settings.particles = particles
settings.output = {'tallies': False}
settings.trigger_active = True
settings.trigger_max_batches = max_batches
# Create an initial uniform spatial source distribution over fissionable zones
bounds = [-10.71, -10.71, -10, 10.71, 10.71, 10.]
uniform_dist = openmc.stats.Box(bounds[:3], bounds[3:], only_fissionable=True)
settings.source = openmc.Source(space=uniform_dist)
# Export to "settings.xml"
settings.export_to_xml()
Let us also create a plot file that we can use to verify that our pin cell geometry was created successfully.
[10]:
# Instantiate a Plot
plot = openmc.Plot(plot_id=1)
plot.filename = 'materials-xy'
plot.origin = [0, 0, 0]
plot.width = [21.5, 21.5]
plot.pixels = [250, 250]
plot.color_by = 'material'
# Show plot
openmc.plot_inline(plot)

As we can see from the plot, we have a nice array of pin cells with fuel, cladding, and water! Before we run our simulation, we need to tell the code what we want to tally. The following code shows how to create a variety of tallies.
[11]:
# Instantiate an empty Tallies object
tallies = openmc.Tallies()
Instantiate a fission rate mesh Tally
[12]:
# Instantiate a tally Mesh
mesh = openmc.RegularMesh(mesh_id=1)
mesh.dimension = [17, 17]
mesh.lower_left = [-10.71, -10.71]
mesh.width = [1.26, 1.26]
# Instantiate tally Filter
mesh_filter = openmc.MeshFilter(mesh)
# Instantiate energy Filter
energy_filter = openmc.EnergyFilter([0, 0.625, 20.0e6])
# Instantiate the Tally
tally = openmc.Tally(name='mesh tally')
tally.filters = [mesh_filter, energy_filter]
tally.scores = ['fission', 'nu-fission']
# Add mesh and Tally to Tallies
tallies.append(tally)
Instantiate a cell Tally with nuclides
[13]:
# Instantiate tally Filter
cell_filter = openmc.CellFilter(fuel_cell)
# Instantiate the tally
tally = openmc.Tally(name='cell tally')
tally.filters = [cell_filter]
tally.scores = ['scatter']
tally.nuclides = ['U235', 'U238']
# Add mesh and tally to Tallies
tallies.append(tally)
Create a “distribcell” Tally. The distribcell filter allows us to tally multiple repeated instances of the same cell throughout the geometry.
[14]:
# Instantiate tally Filter
distribcell_filter = openmc.DistribcellFilter(moderator_cell)
# Instantiate tally Trigger for kicks
trigger = openmc.Trigger(trigger_type='std_dev', threshold=5e-5)
trigger.scores = ['absorption']
# Instantiate the Tally
tally = openmc.Tally(name='distribcell tally')
tally.filters = [distribcell_filter]
tally.scores = ['absorption', 'scatter']
tally.triggers = [trigger]
# Add mesh and tally to Tallies
tallies.append(tally)
[15]:
# Export to "tallies.xml"
tallies.export_to_xml()
Now we a have a complete set of inputs, so we can go ahead and run our simulation.
[16]:
# Remove old HDF5 (summary, statepoint) files
!rm statepoint.*
# Run OpenMC!
openmc.run()
%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
############### %%%%%%%%%%%%%%%%%%%%%%%%
################## %%%%%%%%%%%%%%%%%%%%%%%
################### %%%%%%%%%%%%%%%%%%%%%%%
#################### %%%%%%%%%%%%%%%%%%%%%%
##################### %%%%%%%%%%%%%%%%%%%%%
###################### %%%%%%%%%%%%%%%%%%%%
####################### %%%%%%%%%%%%%%%%%%
####################### %%%%%%%%%%%%%%%%%
###################### %%%%%%%%%%%%%%%%%
#################### %%%%%%%%%%%%%%%%%
################# %%%%%%%%%%%%%%%%%
############### %%%%%%%%%%%%%%%%
############ %%%%%%%%%%%%%%%
######## %%%%%%%%%%%%%%
%%%%%%%%%%%
| The OpenMC Monte Carlo Code
Copyright | 2011-2020 MIT and OpenMC contributors
License | https://docs.openmc.org/en/latest/license.html
Version | 0.12.0
Git SHA1 | 3d90a9f857ec72eae897e054d4225180f1fa4d93
Date/Time | 2020-08-15 07:10:20
OpenMP Threads | 4
Reading settings XML file...
Reading cross sections XML file...
Reading materials XML file...
Reading geometry XML file...
Reading U235 from /home/master/data/nuclear/endfb71_hdf5/U235.h5
Reading U238 from /home/master/data/nuclear/endfb71_hdf5/U238.h5
Reading O16 from /home/master/data/nuclear/endfb71_hdf5/O16.h5
Reading H1 from /home/master/data/nuclear/endfb71_hdf5/H1.h5
Reading B10 from /home/master/data/nuclear/endfb71_hdf5/B10.h5
Reading Zr90 from /home/master/data/nuclear/endfb71_hdf5/Zr90.h5
Minimum neutron data temperature: 294.000000 K
Maximum neutron data temperature: 294.000000 K
Reading tallies XML file...
Preparing distributed cell instances...
Writing summary.h5 file...
Maximum neutron transport energy: 20000000.000000 eV for U235
Initializing source particles...
====================> K EIGENVALUE SIMULATION <====================
Bat./Gen. k Average k
========= ======== ====================
1/1 0.53544
2/1 0.62631
3/1 0.63917
4/1 0.67203
5/1 0.69300
6/1 0.64862
7/1 0.63937 0.64399 +/- 0.00463
8/1 0.67696 0.65498 +/- 0.01131
9/1 0.63216 0.64928 +/- 0.00982
10/1 0.70996 0.66141 +/- 0.01433
11/1 0.69761 0.66745 +/- 0.01316
12/1 0.68662 0.67019 +/- 0.01146
13/1 0.64374 0.66688 +/- 0.01046
14/1 0.69121 0.66958 +/- 0.00961
15/1 0.72125 0.67475 +/- 0.01003
16/1 0.72706 0.67950 +/- 0.01024
17/1 0.69623 0.68090 +/- 0.00945
18/1 0.70953 0.68310 +/- 0.00897
19/1 0.69026 0.68361 +/- 0.00832
20/1 0.68633 0.68379 +/- 0.00775
Triggers unsatisfied, max unc./thresh. is 75.24758750489383 for absorption in
tally 3
WARNING: The estimated number of batches is 84938 --- greater than max batches
Creating state point statepoint.020.h5...
21/1 0.68310 0.68375 +/- 0.00725
Triggers unsatisfied, max unc./thresh. is 71.20148627325992 for absorption in
tally 3
WARNING: The estimated number of batches is 81120 --- greater than max batches
22/1 0.68679 0.68393 +/- 0.00681
Triggers unsatisfied, max unc./thresh. is 66.94650483064697 for absorption in
tally 3
WARNING: The estimated number of batches is 76197 --- greater than max batches
23/1 0.67440 0.68340 +/- 0.00644
Triggers unsatisfied, max unc./thresh. is 63.553590826021285 for absorption in
tally 3
WARNING: The estimated number of batches is 72709 --- greater than max batches
24/1 0.67483 0.68295 +/- 0.00611
Triggers unsatisfied, max unc./thresh. is 60.37873858685279 for absorption in
tally 3
WARNING: The estimated number of batches is 69272 --- greater than max batches
25/1 0.71558 0.68458 +/- 0.00602
Triggers unsatisfied, max unc./thresh. is 60.34535216026281 for absorption in
tally 3
WARNING: The estimated number of batches is 72837 --- greater than max batches
26/1 0.71853 0.68620 +/- 0.00595
Triggers unsatisfied, max unc./thresh. is 59.60875760463032 for absorption in
tally 3
WARNING: The estimated number of batches is 74623 --- greater than max batches
27/1 0.67455 0.68567 +/- 0.00570
Triggers unsatisfied, max unc./thresh. is 57.228951643423976 for absorption in
tally 3
WARNING: The estimated number of batches is 72059 --- greater than max batches
28/1 0.69435 0.68605 +/- 0.00546
Triggers unsatisfied, max unc./thresh. is 56.194065573871285 for absorption in
tally 3
WARNING: The estimated number of batches is 72634 --- greater than max batches
29/1 0.67706 0.68567 +/- 0.00524
Triggers unsatisfied, max unc./thresh. is 53.86022066923874 for absorption in
tally 3
WARNING: The estimated number of batches is 69628 --- greater than max batches
30/1 0.69294 0.68596 +/- 0.00504
Triggers unsatisfied, max unc./thresh. is 51.73413206858763 for absorption in
tally 3
WARNING: The estimated number of batches is 66916 --- greater than max batches
31/1 0.69108 0.68616 +/- 0.00484
Triggers unsatisfied, max unc./thresh. is 49.71174462484801 for absorption in
tally 3
WARNING: The estimated number of batches is 64258 --- greater than max batches
32/1 0.68089 0.68596 +/- 0.00466
Triggers unsatisfied, max unc./thresh. is 50.80993117627794 for absorption in
tally 3
WARNING: The estimated number of batches is 69710 --- greater than max batches
33/1 0.67698 0.68564 +/- 0.00450
Triggers unsatisfied, max unc./thresh. is 50.46659333785448 for absorption in
tally 3
WARNING: The estimated number of batches is 71318 --- greater than max batches
34/1 0.68167 0.68551 +/- 0.00435
Triggers unsatisfied, max unc./thresh. is 48.852656603250665 for absorption in
tally 3
WARNING: The estimated number of batches is 69216 --- greater than max batches
35/1 0.67760 0.68524 +/- 0.00421
Triggers unsatisfied, max unc./thresh. is 48.5685583427197 for absorption in
tally 3
WARNING: The estimated number of batches is 70773 --- greater than max batches
36/1 0.67628 0.68495 +/- 0.00408
Triggers unsatisfied, max unc./thresh. is 47.77661998216646 for absorption in
tally 3
WARNING: The estimated number of batches is 70766 --- greater than max batches
37/1 0.66736 0.68440 +/- 0.00399
Triggers unsatisfied, max unc./thresh. is 46.57810773879176 for absorption in
tally 3
WARNING: The estimated number of batches is 69430 --- greater than max batches
38/1 0.71026 0.68519 +/- 0.00395
Triggers unsatisfied, max unc./thresh. is 45.876107560616674 for absorption in
tally 3
WARNING: The estimated number of batches is 69458 --- greater than max batches
39/1 0.67674 0.68494 +/- 0.00384
Triggers unsatisfied, max unc./thresh. is 45.30341918376721 for absorption in
tally 3
WARNING: The estimated number of batches is 69787 --- greater than max batches
40/1 0.69360 0.68519 +/- 0.00373
Triggers unsatisfied, max unc./thresh. is 44.018056562863386 for absorption in
tally 3
WARNING: The estimated number of batches is 67821 --- greater than max batches
41/1 0.70987 0.68587 +/- 0.00369
Triggers unsatisfied, max unc./thresh. is 42.781078099052785 for absorption in
tally 3
WARNING: The estimated number of batches is 65893 --- greater than max batches
42/1 0.68780 0.68592 +/- 0.00359
Triggers unsatisfied, max unc./thresh. is 41.60877069209228 for absorption in
tally 3
WARNING: The estimated number of batches is 64063 --- greater than max batches
43/1 0.69223 0.68609 +/- 0.00350
Triggers unsatisfied, max unc./thresh. is 42.44932759168626 for absorption in
tally 3
WARNING: The estimated number of batches is 68479 --- greater than max batches
44/1 0.69561 0.68633 +/- 0.00342
Triggers unsatisfied, max unc./thresh. is 41.34743776753899 for absorption in
tally 3
WARNING: The estimated number of batches is 66680 --- greater than max batches
45/1 0.67503 0.68605 +/- 0.00334
Triggers unsatisfied, max unc./thresh. is 40.97332186124358 for absorption in
tally 3
WARNING: The estimated number of batches is 67158 --- greater than max batches
46/1 0.67290 0.68573 +/- 0.00328
Triggers unsatisfied, max unc./thresh. is 40.24678448931756 for absorption in
tally 3
WARNING: The estimated number of batches is 66417 --- greater than max batches
47/1 0.67355 0.68544 +/- 0.00321
Triggers unsatisfied, max unc./thresh. is 40.42620640829592 for absorption in
tally 3
WARNING: The estimated number of batches is 68645 --- greater than max batches
48/1 0.71383 0.68610 +/- 0.00320
Triggers unsatisfied, max unc./thresh. is 39.90662320308606 for absorption in
tally 3
WARNING: The estimated number of batches is 68485 --- greater than max batches
49/1 0.68389 0.68605 +/- 0.00313
Triggers unsatisfied, max unc./thresh. is 39.075369568753906 for absorption in
tally 3
WARNING: The estimated number of batches is 67188 --- greater than max batches
50/1 0.73148 0.68706 +/- 0.00322
Triggers unsatisfied, max unc./thresh. is 38.57054567218653 for absorption in
tally 3
WARNING: The estimated number of batches is 66951 --- greater than max batches
51/1 0.69796 0.68730 +/- 0.00316
Triggers unsatisfied, max unc./thresh. is 38.21572703507316 for absorption in
tally 3
WARNING: The estimated number of batches is 67186 --- greater than max batches
52/1 0.70691 0.68771 +/- 0.00312
Triggers unsatisfied, max unc./thresh. is 37.50971908717773 for absorption in
tally 3
WARNING: The estimated number of batches is 66134 --- greater than max batches
53/1 0.69104 0.68778 +/- 0.00306
Triggers unsatisfied, max unc./thresh. is 36.824732312223716 for absorption in
tally 3
WARNING: The estimated number of batches is 65096 --- greater than max batches
54/1 0.74368 0.68892 +/- 0.00320
Triggers unsatisfied, max unc./thresh. is 36.20814737643575 for absorption in
tally 3
WARNING: The estimated number of batches is 64246 --- greater than max batches
55/1 0.67371 0.68862 +/- 0.00315
Triggers unsatisfied, max unc./thresh. is 35.48607231512293 for absorption in
tally 3
WARNING: The estimated number of batches is 62969 --- greater than max batches
56/1 0.67846 0.68842 +/- 0.00310
Triggers unsatisfied, max unc./thresh. is 35.34421893287461 for absorption in
tally 3
WARNING: The estimated number of batches is 63715 --- greater than max batches
57/1 0.66351 0.68794 +/- 0.00307
Triggers unsatisfied, max unc./thresh. is 34.67062652878957 for absorption in
tally 3
WARNING: The estimated number of batches is 62512 --- greater than max batches
58/1 0.67049 0.68761 +/- 0.00303
Triggers unsatisfied, max unc./thresh. is 34.22135922543247 for absorption in
tally 3
WARNING: The estimated number of batches is 62074 --- greater than max batches
59/1 0.66967 0.68728 +/- 0.00299
Triggers unsatisfied, max unc./thresh. is 33.66881484408945 for absorption in
tally 3
WARNING: The estimated number of batches is 61219 --- greater than max batches
60/1 0.70271 0.68756 +/- 0.00295
Triggers unsatisfied, max unc./thresh. is 33.19914799505717 for absorption in
tally 3
WARNING: The estimated number of batches is 60626 --- greater than max batches
61/1 0.70035 0.68779 +/- 0.00291
Triggers unsatisfied, max unc./thresh. is 32.65594936729897 for absorption in
tally 3
WARNING: The estimated number of batches is 59725 --- greater than max batches
62/1 0.66274 0.68735 +/- 0.00289
Triggers unsatisfied, max unc./thresh. is 32.15622046485561 for absorption in
tally 3
WARNING: The estimated number of batches is 58945 --- greater than max batches
63/1 0.68607 0.68733 +/- 0.00284
Triggers unsatisfied, max unc./thresh. is 31.601225649282494 for absorption in
tally 3
WARNING: The estimated number of batches is 57926 --- greater than max batches
64/1 0.66518 0.68695 +/- 0.00282
Triggers unsatisfied, max unc./thresh. is 31.12129365572805 for absorption in
tally 3
WARNING: The estimated number of batches is 57149 --- greater than max batches
65/1 0.65999 0.68650 +/- 0.00281
Triggers unsatisfied, max unc./thresh. is 30.641988019531464 for absorption in
tally 3
WARNING: The estimated number of batches is 56341 --- greater than max batches
66/1 0.67843 0.68637 +/- 0.00276
Triggers unsatisfied, max unc./thresh. is 30.320463443580458 for absorption in
tally 3
WARNING: The estimated number of batches is 56085 --- greater than max batches
67/1 0.69295 0.68648 +/- 0.00272
Triggers unsatisfied, max unc./thresh. is 30.06051080038397 for absorption in
tally 3
WARNING: The estimated number of batches is 56031 --- greater than max batches
68/1 0.69158 0.68656 +/- 0.00268
Triggers unsatisfied, max unc./thresh. is 29.7400907913873 for absorption in
tally 3
WARNING: The estimated number of batches is 55727 --- greater than max batches
69/1 0.69825 0.68674 +/- 0.00264
Triggers unsatisfied, max unc./thresh. is 29.278619659445805 for absorption in
tally 3
WARNING: The estimated number of batches is 54869 --- greater than max batches
70/1 0.73637 0.68750 +/- 0.00271
Triggers unsatisfied, max unc./thresh. is 28.945044018568716 for absorption in
tally 3
WARNING: The estimated number of batches is 54464 --- greater than max batches
71/1 0.64301 0.68683 +/- 0.00275
Triggers unsatisfied, max unc./thresh. is 28.73677928804667 for absorption in
tally 3
WARNING: The estimated number of batches is 54508 --- greater than max batches
72/1 0.71506 0.68725 +/- 0.00274
Triggers unsatisfied, max unc./thresh. is 29.20796537704291 for absorption in
tally 3
WARNING: The estimated number of batches is 57164 --- greater than max batches
73/1 0.69203 0.68732 +/- 0.00270
Triggers unsatisfied, max unc./thresh. is 29.56297016014581 for absorption in
tally 3
WARNING: The estimated number of batches is 59435 --- greater than max batches
74/1 0.69208 0.68739 +/- 0.00267
Triggers unsatisfied, max unc./thresh. is 29.545241442413783 for absorption in
tally 3
WARNING: The estimated number of batches is 60237 --- greater than max batches
75/1 0.65717 0.68696 +/- 0.00266
Triggers unsatisfied, max unc./thresh. is 29.284013224166248 for absorption in
tally 3
WARNING: The estimated number of batches is 60034 --- greater than max batches
76/1 0.70992 0.68728 +/- 0.00265
Triggers unsatisfied, max unc./thresh. is 29.00034584995327 for absorption in
tally 3
WARNING: The estimated number of batches is 59718 --- greater than max batches
77/1 0.65590 0.68685 +/- 0.00264
Triggers unsatisfied, max unc./thresh. is 28.867967845905174 for absorption in
tally 3
WARNING: The estimated number of batches is 60007 --- greater than max batches
78/1 0.64439 0.68626 +/- 0.00267
Triggers unsatisfied, max unc./thresh. is 29.016012595317935 for absorption in
tally 3
WARNING: The estimated number of batches is 61466 --- greater than max batches
79/1 0.66295 0.68595 +/- 0.00265
Triggers unsatisfied, max unc./thresh. is 28.626245464278142 for absorption in
tally 3
WARNING: The estimated number of batches is 60646 --- greater than max batches
80/1 0.66672 0.68569 +/- 0.00263
Triggers unsatisfied, max unc./thresh. is 28.24218910063624 for absorption in
tally 3
WARNING: The estimated number of batches is 59827 --- greater than max batches
81/1 0.69110 0.68576 +/- 0.00260
Triggers unsatisfied, max unc./thresh. is 27.917349908027763 for absorption in
tally 3
WARNING: The estimated number of batches is 59238 --- greater than max batches
82/1 0.67481 0.68562 +/- 0.00257
Triggers unsatisfied, max unc./thresh. is 28.01946018168837 for absorption in
tally 3
WARNING: The estimated number of batches is 60457 --- greater than max batches
83/1 0.72216 0.68609 +/- 0.00258
Triggers unsatisfied, max unc./thresh. is 27.931394620754766 for absorption in
tally 3
WARNING: The estimated number of batches is 60858 --- greater than max batches
84/1 0.68429 0.68607 +/- 0.00254
Triggers unsatisfied, max unc./thresh. is 27.713137470531738 for absorption in
tally 3
WARNING: The estimated number of batches is 60679 --- greater than max batches
85/1 0.65458 0.68567 +/- 0.00254
Triggers unsatisfied, max unc./thresh. is 27.364539968246927 for absorption in
tally 3
WARNING: The estimated number of batches is 59911 --- greater than max batches
86/1 0.69966 0.68585 +/- 0.00252
Triggers unsatisfied, max unc./thresh. is 27.178113974435043 for absorption in
tally 3
WARNING: The estimated number of batches is 59836 --- greater than max batches
87/1 0.64776 0.68538 +/- 0.00253
Triggers unsatisfied, max unc./thresh. is 26.941566345072534 for absorption in
tally 3
WARNING: The estimated number of batches is 59525 --- greater than max batches
88/1 0.62737 0.68468 +/- 0.00259
Triggers unsatisfied, max unc./thresh. is 26.73959660667411 for absorption in
tally 3
WARNING: The estimated number of batches is 59351 --- greater than max batches
89/1 0.69779 0.68484 +/- 0.00257
Triggers unsatisfied, max unc./thresh. is 26.490234865810894 for absorption in
tally 3
WARNING: The estimated number of batches is 58951 --- greater than max batches
90/1 0.67312 0.68470 +/- 0.00254
Triggers unsatisfied, max unc./thresh. is 26.24036001465229 for absorption in
tally 3
WARNING: The estimated number of batches is 58533 --- greater than max batches
91/1 0.69289 0.68480 +/- 0.00251
Triggers unsatisfied, max unc./thresh. is 25.936795778335345 for absorption in
tally 3
WARNING: The estimated number of batches is 57859 --- greater than max batches
92/1 0.69884 0.68496 +/- 0.00249
Triggers unsatisfied, max unc./thresh. is 25.695465963215582 for absorption in
tally 3
WARNING: The estimated number of batches is 57448 --- greater than max batches
93/1 0.71351 0.68528 +/- 0.00248
Triggers unsatisfied, max unc./thresh. is 25.49001212499821 for absorption in
tally 3
WARNING: The estimated number of batches is 57183 --- greater than max batches
94/1 0.65602 0.68495 +/- 0.00248
Triggers unsatisfied, max unc./thresh. is 25.350859463183905 for absorption in
tally 3
WARNING: The estimated number of batches is 57203 --- greater than max batches
95/1 0.72223 0.68537 +/- 0.00248
Triggers unsatisfied, max unc./thresh. is 25.157803279393637 for absorption in
tally 3
WARNING: The estimated number of batches is 56968 --- greater than max batches
96/1 0.67930 0.68530 +/- 0.00246
Triggers unsatisfied, max unc./thresh. is 24.92205849077747 for absorption in
tally 3
WARNING: The estimated number of batches is 56526 --- greater than max batches
97/1 0.66201 0.68505 +/- 0.00244
Triggers unsatisfied, max unc./thresh. is 24.653967027285237 for absorption in
tally 3
WARNING: The estimated number of batches is 55925 --- greater than max batches
98/1 0.71110 0.68533 +/- 0.00243
Triggers unsatisfied, max unc./thresh. is 24.566957281211884 for absorption in
tally 3
WARNING: The estimated number of batches is 56134 --- greater than max batches
99/1 0.69409 0.68542 +/- 0.00241
Triggers unsatisfied, max unc./thresh. is 24.581943149247135 for absorption in
tally 3
WARNING: The estimated number of batches is 56807 --- greater than max batches
100/1 0.71197 0.68570 +/- 0.00240
Triggers unsatisfied, max unc./thresh. is 24.444112571967217 for absorption in
tally 3
WARNING: The estimated number of batches is 56769 --- greater than max batches
101/1 0.71713 0.68603 +/- 0.00240
Triggers unsatisfied, max unc./thresh. is 24.18957776896016 for absorption in
tally 3
WARNING: The estimated number of batches is 56179 --- greater than max batches
102/1 0.68143 0.68598 +/- 0.00237
Triggers unsatisfied, max unc./thresh. is 23.97797098066553 for absorption in
tally 3
WARNING: The estimated number of batches is 55775 --- greater than max batches
103/1 0.69936 0.68612 +/- 0.00235
Triggers unsatisfied, max unc./thresh. is 24.253000406602812 for absorption in
tally 3
WARNING: The estimated number of batches is 57650 --- greater than max batches
104/1 0.65247 0.68578 +/- 0.00235
Triggers unsatisfied, max unc./thresh. is 24.593482483379837 for absorption in
tally 3
WARNING: The estimated number of batches is 59885 --- greater than max batches
105/1 0.66517 0.68557 +/- 0.00234
Triggers unsatisfied, max unc./thresh. is 24.37904760701804 for absorption in
tally 3
WARNING: The estimated number of batches is 59439 --- greater than max batches
106/1 0.67814 0.68550 +/- 0.00232
Triggers unsatisfied, max unc./thresh. is 24.142311084988883 for absorption in
tally 3
WARNING: The estimated number of batches is 58873 --- greater than max batches
107/1 0.67788 0.68542 +/- 0.00229
Triggers unsatisfied, max unc./thresh. is 23.935477435724106 for absorption in
tally 3
WARNING: The estimated number of batches is 58442 --- greater than max batches
108/1 0.68016 0.68537 +/- 0.00227
Triggers unsatisfied, max unc./thresh. is 24.532504688648594 for absorption in
tally 3
WARNING: The estimated number of batches is 61995 --- greater than max batches
109/1 0.66963 0.68522 +/- 0.00226
Triggers unsatisfied, max unc./thresh. is 24.354532539671386 for absorption in
tally 3
WARNING: The estimated number of batches is 61692 --- greater than max batches
110/1 0.67556 0.68513 +/- 0.00224
Triggers unsatisfied, max unc./thresh. is 24.16165322902175 for absorption in
tally 3
WARNING: The estimated number of batches is 61303 --- greater than max batches
111/1 0.68273 0.68511 +/- 0.00222
Triggers unsatisfied, max unc./thresh. is 24.00069508298176 for absorption in
tally 3
WARNING: The estimated number of batches is 61065 --- greater than max batches
112/1 0.69505 0.68520 +/- 0.00220
Triggers unsatisfied, max unc./thresh. is 23.791656279909404 for absorption in
tally 3
WARNING: The estimated number of batches is 60572 --- greater than max batches
113/1 0.69385 0.68528 +/- 0.00218
Triggers unsatisfied, max unc./thresh. is 23.667941020219764 for absorption in
tally 3
WARNING: The estimated number of batches is 60504 --- greater than max batches
114/1 0.65352 0.68499 +/- 0.00218
Triggers unsatisfied, max unc./thresh. is 23.469658485546123 for absorption in
tally 3
WARNING: The estimated number of batches is 60045 --- greater than max batches
115/1 0.68339 0.68497 +/- 0.00216
Triggers unsatisfied, max unc./thresh. is 23.259123161328624 for absorption in
tally 3
WARNING: The estimated number of batches is 59514 --- greater than max batches
116/1 0.65854 0.68474 +/- 0.00215
Triggers unsatisfied, max unc./thresh. is 23.06250977653337 for absorption in
tally 3
WARNING: The estimated number of batches is 59044 --- greater than max batches
117/1 0.66907 0.68460 +/- 0.00214
Triggers unsatisfied, max unc./thresh. is 22.874382198219536 for absorption in
tally 3
WARNING: The estimated number of batches is 58608 --- greater than max batches
118/1 0.68165 0.68457 +/- 0.00212
Triggers unsatisfied, max unc./thresh. is 22.709602691165983 for absorption in
tally 3
WARNING: The estimated number of batches is 58283 --- greater than max batches
119/1 0.70967 0.68479 +/- 0.00211
Triggers unsatisfied, max unc./thresh. is 22.509869996658225 for absorption in
tally 3
WARNING: The estimated number of batches is 57769 --- greater than max batches
120/1 0.65543 0.68453 +/- 0.00211
Triggers unsatisfied, max unc./thresh. is 22.422104322874098 for absorption in
tally 3
WARNING: The estimated number of batches is 57822 --- greater than max batches
121/1 0.67305 0.68444 +/- 0.00209
Triggers unsatisfied, max unc./thresh. is 22.32834321567902 for absorption in
tally 3
WARNING: The estimated number of batches is 57838 --- greater than max batches
122/1 0.68206 0.68441 +/- 0.00207
Triggers unsatisfied, max unc./thresh. is 22.155196965032374 for absorption in
tally 3
WARNING: The estimated number of batches is 57435 --- greater than max batches
123/1 0.71125 0.68464 +/- 0.00207
Triggers unsatisfied, max unc./thresh. is 21.96683678913398 for absorption in
tally 3
WARNING: The estimated number of batches is 56945 --- greater than max batches
124/1 0.65918 0.68443 +/- 0.00206
Triggers unsatisfied, max unc./thresh. is 21.78216358933223 for absorption in
tally 3
WARNING: The estimated number of batches is 56467 --- greater than max batches
125/1 0.68122 0.68440 +/- 0.00205
Triggers unsatisfied, max unc./thresh. is 21.681928420505304 for absorption in
tally 3
WARNING: The estimated number of batches is 56418 --- greater than max batches
126/1 0.66900 0.68427 +/- 0.00203
Triggers unsatisfied, max unc./thresh. is 21.60631058168512 for absorption in
tally 3
WARNING: The estimated number of batches is 56492 --- greater than max batches
127/1 0.66742 0.68414 +/- 0.00202
Triggers unsatisfied, max unc./thresh. is 21.468291123480988 for absorption in
tally 3
WARNING: The estimated number of batches is 56234 --- greater than max batches
128/1 0.66971 0.68402 +/- 0.00201
Triggers unsatisfied, max unc./thresh. is 21.313238544974386 for absorption in
tally 3
WARNING: The estimated number of batches is 55879 --- greater than max batches
129/1 0.68183 0.68400 +/- 0.00199
Triggers unsatisfied, max unc./thresh. is 21.314008888585132 for absorption in
tally 3
WARNING: The estimated number of batches is 56337 --- greater than max batches
130/1 0.68403 0.68400 +/- 0.00197
Triggers unsatisfied, max unc./thresh. is 21.159444482258046 for absorption in
tally 3
WARNING: The estimated number of batches is 55971 --- greater than max batches
131/1 0.69137 0.68406 +/- 0.00196
Triggers unsatisfied, max unc./thresh. is 21.24931160989673 for absorption in
tally 3
WARNING: The estimated number of batches is 56899 --- greater than max batches
132/1 0.67481 0.68399 +/- 0.00195
Triggers unsatisfied, max unc./thresh. is 21.164512281281944 for absorption in
tally 3
WARNING: The estimated number of batches is 56893 --- greater than max batches
133/1 0.70390 0.68414 +/- 0.00194
Triggers unsatisfied, max unc./thresh. is 21.084176856795946 for absorption in
tally 3
WARNING: The estimated number of batches is 56907 --- greater than max batches
134/1 0.67961 0.68411 +/- 0.00192
Triggers unsatisfied, max unc./thresh. is 21.48684646255342 for absorption in
tally 3
WARNING: The estimated number of batches is 59563 --- greater than max batches
135/1 0.65362 0.68387 +/- 0.00192
Triggers unsatisfied, max unc./thresh. is 21.41235779526348 for absorption in
tally 3
WARNING: The estimated number of batches is 59609 --- greater than max batches
136/1 0.63946 0.68353 +/- 0.00194
Triggers unsatisfied, max unc./thresh. is 21.30299014546295 for absorption in
tally 3
WARNING: The estimated number of batches is 59456 --- greater than max batches
137/1 0.64818 0.68327 +/- 0.00194
Triggers unsatisfied, max unc./thresh. is 21.159761745415484 for absorption in
tally 3
WARNING: The estimated number of batches is 59107 --- greater than max batches
138/1 0.68975 0.68331 +/- 0.00193
Triggers unsatisfied, max unc./thresh. is 21.000094566393475 for absorption in
tally 3
WARNING: The estimated number of batches is 58659 --- greater than max batches
139/1 0.67280 0.68324 +/- 0.00191
Triggers unsatisfied, max unc./thresh. is 20.853656171297644 for absorption in
tally 3
WARNING: The estimated number of batches is 58279 --- greater than max batches
140/1 0.66857 0.68313 +/- 0.00190
Triggers unsatisfied, max unc./thresh. is 20.709591767033707 for absorption in
tally 3
WARNING: The estimated number of batches is 57905 --- greater than max batches
141/1 0.68175 0.68312 +/- 0.00189
Triggers unsatisfied, max unc./thresh. is 20.560813068689416 for absorption in
tally 3
WARNING: The estimated number of batches is 57499 --- greater than max batches
142/1 0.72210 0.68340 +/- 0.00190
Triggers unsatisfied, max unc./thresh. is 20.54814917791921 for absorption in
tally 3
WARNING: The estimated number of batches is 57851 --- greater than max batches
143/1 0.67361 0.68333 +/- 0.00188
Triggers unsatisfied, max unc./thresh. is 20.4177880049802 for absorption in
tally 3
WARNING: The estimated number of batches is 57536 --- greater than max batches
144/1 0.65862 0.68315 +/- 0.00188
Triggers unsatisfied, max unc./thresh. is 21.229890183572195 for absorption in
tally 3
WARNING: The estimated number of batches is 62654 --- greater than max batches
145/1 0.69713 0.68325 +/- 0.00187
Triggers unsatisfied, max unc./thresh. is 21.34513800240435 for absorption in
tally 3
WARNING: The estimated number of batches is 63792 --- greater than max batches
146/1 0.72980 0.68358 +/- 0.00188
Triggers unsatisfied, max unc./thresh. is 21.60165210412777 for absorption in
tally 3
WARNING: The estimated number of batches is 65801 --- greater than max batches
147/1 0.70004 0.68370 +/- 0.00187
Triggers unsatisfied, max unc./thresh. is 21.596734424310384 for absorption in
tally 3
WARNING: The estimated number of batches is 66237 --- greater than max batches
148/1 0.68882 0.68374 +/- 0.00186
Triggers unsatisfied, max unc./thresh. is 21.447240534346236 for absorption in
tally 3
WARNING: The estimated number of batches is 65783 --- greater than max batches
149/1 0.70401 0.68388 +/- 0.00185
Triggers unsatisfied, max unc./thresh. is 21.424993974056104 for absorption in
tally 3
WARNING: The estimated number of batches is 66106 --- greater than max batches
150/1 0.72110 0.68413 +/- 0.00186
Triggers unsatisfied, max unc./thresh. is 21.27792348945665 for absorption in
tally 3
WARNING: The estimated number of batches is 65654 --- greater than max batches
151/1 0.65918 0.68396 +/- 0.00185
Triggers unsatisfied, max unc./thresh. is 21.378637401006184 for absorption in
tally 3
WARNING: The estimated number of batches is 66734 --- greater than max batches
152/1 0.67751 0.68392 +/- 0.00184
Triggers unsatisfied, max unc./thresh. is 21.25974745003047 for absorption in
tally 3
WARNING: The estimated number of batches is 66446 --- greater than max batches
153/1 0.69302 0.68398 +/- 0.00183
Triggers unsatisfied, max unc./thresh. is 21.16055371271148 for absorption in
tally 3
WARNING: The estimated number of batches is 66275 --- greater than max batches
154/1 0.67102 0.68389 +/- 0.00182
Triggers unsatisfied, max unc./thresh. is 21.0227808264386 for absorption in
tally 3
WARNING: The estimated number of batches is 65857 --- greater than max batches
155/1 0.64427 0.68363 +/- 0.00183
Triggers unsatisfied, max unc./thresh. is 20.882547322553506 for absorption in
tally 3
WARNING: The estimated number of batches is 65418 --- greater than max batches
156/1 0.68488 0.68364 +/- 0.00181
Triggers unsatisfied, max unc./thresh. is 20.797424126850476 for absorption in
tally 3
WARNING: The estimated number of batches is 65318 --- greater than max batches
157/1 0.67337 0.68357 +/- 0.00180
Triggers unsatisfied, max unc./thresh. is 20.67015745584828 for absorption in
tally 3
WARNING: The estimated number of batches is 64948 --- greater than max batches
158/1 0.66662 0.68346 +/- 0.00180
Triggers unsatisfied, max unc./thresh. is 20.568519956722266 for absorption in
tally 3
WARNING: The estimated number of batches is 64734 --- greater than max batches
159/1 0.62697 0.68309 +/- 0.00182
Triggers unsatisfied, max unc./thresh. is 20.47085213159483 for absorption in
tally 3
WARNING: The estimated number of batches is 64540 --- greater than max batches
160/1 0.68300 0.68309 +/- 0.00181
Triggers unsatisfied, max unc./thresh. is 20.34586209866351 for absorption in
tally 3
WARNING: The estimated number of batches is 64168 --- greater than max batches
161/1 0.68918 0.68313 +/- 0.00180
Triggers unsatisfied, max unc./thresh. is 20.23505377614212 for absorption in
tally 3
WARNING: The estimated number of batches is 63881 --- greater than max batches
162/1 0.70939 0.68330 +/- 0.00179
Triggers unsatisfied, max unc./thresh. is 20.21114215977674 for absorption in
tally 3
WARNING: The estimated number of batches is 64138 --- greater than max batches
163/1 0.69681 0.68338 +/- 0.00179
Triggers unsatisfied, max unc./thresh. is 20.163170350438893 for absorption in
tally 3
WARNING: The estimated number of batches is 64241 --- greater than max batches
164/1 0.66454 0.68326 +/- 0.00178
Triggers unsatisfied, max unc./thresh. is 20.109882525638955 for absorption in
tally 3
WARNING: The estimated number of batches is 64306 --- greater than max batches
165/1 0.68804 0.68329 +/- 0.00177
Triggers unsatisfied, max unc./thresh. is 20.033653897136055 for absorption in
tally 3
WARNING: The estimated number of batches is 64221 --- greater than max batches
166/1 0.66078 0.68315 +/- 0.00176
Triggers unsatisfied, max unc./thresh. is 19.916131324861123 for absorption in
tally 3
WARNING: The estimated number of batches is 63867 --- greater than max batches
167/1 0.65762 0.68300 +/- 0.00176
Triggers unsatisfied, max unc./thresh. is 19.85097950868946 for absorption in
tally 3
WARNING: The estimated number of batches is 63843 --- greater than max batches
168/1 0.69267 0.68306 +/- 0.00175
Triggers unsatisfied, max unc./thresh. is 19.729436003984436 for absorption in
tally 3
WARNING: The estimated number of batches is 63453 --- greater than max batches
169/1 0.67859 0.68303 +/- 0.00174
Triggers unsatisfied, max unc./thresh. is 19.61178427698242 for absorption in
tally 3
WARNING: The estimated number of batches is 63084 --- greater than max batches
170/1 0.66545 0.68292 +/- 0.00173
Triggers unsatisfied, max unc./thresh. is 19.495197332905757 for absorption in
tally 3
WARNING: The estimated number of batches is 62716 --- greater than max batches
171/1 0.66716 0.68283 +/- 0.00172
Triggers unsatisfied, max unc./thresh. is 19.47044861415963 for absorption in
tally 3
WARNING: The estimated number of batches is 62936 --- greater than max batches
172/1 0.70008 0.68293 +/- 0.00172
Triggers unsatisfied, max unc./thresh. is 19.382801191970024 for absorption in
tally 3
WARNING: The estimated number of batches is 62746 --- greater than max batches
173/1 0.69417 0.68300 +/- 0.00171
Triggers unsatisfied, max unc./thresh. is 19.270038663528165 for absorption in
tally 3
WARNING: The estimated number of batches is 62390 --- greater than max batches
174/1 0.66458 0.68289 +/- 0.00170
Triggers unsatisfied, max unc./thresh. is 19.281533726364312 for absorption in
tally 3
WARNING: The estimated number of batches is 62836 --- greater than max batches
175/1 0.65867 0.68275 +/- 0.00170
Triggers unsatisfied, max unc./thresh. is 19.234847030404104 for absorption in
tally 3
WARNING: The estimated number of batches is 62902 --- greater than max batches
176/1 0.69631 0.68283 +/- 0.00169
Triggers unsatisfied, max unc./thresh. is 19.13381099709457 for absorption in
tally 3
WARNING: The estimated number of batches is 62609 --- greater than max batches
177/1 0.71142 0.68299 +/- 0.00169
Triggers unsatisfied, max unc./thresh. is 19.022563643493143 for absorption in
tally 3
WARNING: The estimated number of batches is 62245 --- greater than max batches
178/1 0.68640 0.68301 +/- 0.00168
Triggers unsatisfied, max unc./thresh. is 19.03176453708651 for absorption in
tally 3
WARNING: The estimated number of batches is 62667 --- greater than max batches
179/1 0.70448 0.68313 +/- 0.00167
Triggers unsatisfied, max unc./thresh. is 19.088395456136563 for absorption in
tally 3
WARNING: The estimated number of batches is 63405 --- greater than max batches
180/1 0.70538 0.68326 +/- 0.00167
Triggers unsatisfied, max unc./thresh. is 18.98864751831452 for absorption in
tally 3
WARNING: The estimated number of batches is 63105 --- greater than max batches
181/1 0.65591 0.68311 +/- 0.00166
Triggers unsatisfied, max unc./thresh. is 18.911017891051518 for absorption in
tally 3
WARNING: The estimated number of batches is 62948 --- greater than max batches
182/1 0.72818 0.68336 +/- 0.00167
Triggers unsatisfied, max unc./thresh. is 18.808510226366458 for absorption in
tally 3
WARNING: The estimated number of batches is 62621 --- greater than max batches
183/1 0.67896 0.68334 +/- 0.00167
Triggers unsatisfied, max unc./thresh. is 18.825142861337717 for absorption in
tally 3
WARNING: The estimated number of batches is 63086 --- greater than max batches
184/1 0.65442 0.68317 +/- 0.00166
Triggers unsatisfied, max unc./thresh. is 18.79514029258707 for absorption in
tally 3
WARNING: The estimated number of batches is 63239 --- greater than max batches
185/1 0.68885 0.68321 +/- 0.00165
Triggers unsatisfied, max unc./thresh. is 18.76276176864163 for absorption in
tally 3
WARNING: The estimated number of batches is 63373 --- greater than max batches
186/1 0.68893 0.68324 +/- 0.00165
Triggers unsatisfied, max unc./thresh. is 18.690155368597363 for absorption in
tally 3
WARNING: The estimated number of batches is 63233 --- greater than max batches
187/1 0.68918 0.68327 +/- 0.00164
Triggers unsatisfied, max unc./thresh. is 18.590144288270153 for absorption in
tally 3
WARNING: The estimated number of batches is 62904 --- greater than max batches
188/1 0.69854 0.68335 +/- 0.00163
Triggers unsatisfied, max unc./thresh. is 18.61460656150607 for absorption in
tally 3
WARNING: The estimated number of batches is 63416 --- greater than max batches
189/1 0.66324 0.68324 +/- 0.00162
Triggers unsatisfied, max unc./thresh. is 18.518608099237504 for absorption in
tally 3
WARNING: The estimated number of batches is 63106 --- greater than max batches
190/1 0.69450 0.68331 +/- 0.00162
Triggers unsatisfied, max unc./thresh. is 18.425351661292233 for absorption in
tally 3
WARNING: The estimated number of batches is 62812 --- greater than max batches
191/1 0.68953 0.68334 +/- 0.00161
Triggers unsatisfied, max unc./thresh. is 18.328779429843646 for absorption in
tally 3
WARNING: The estimated number of batches is 62491 --- greater than max batches
192/1 0.66621 0.68325 +/- 0.00160
Triggers unsatisfied, max unc./thresh. is 18.28094973389564 for absorption in
tally 3
WARNING: The estimated number of batches is 62500 --- greater than max batches
193/1 0.71102 0.68339 +/- 0.00160
Triggers unsatisfied, max unc./thresh. is 18.19949730061142 for absorption in
tally 3
WARNING: The estimated number of batches is 62275 --- greater than max batches
194/1 0.65341 0.68324 +/- 0.00160
Triggers unsatisfied, max unc./thresh. is 18.159054737369345 for absorption in
tally 3
WARNING: The estimated number of batches is 62328 --- greater than max batches
195/1 0.70061 0.68333 +/- 0.00159
Triggers unsatisfied, max unc./thresh. is 18.082465954324594 for absorption in
tally 3
WARNING: The estimated number of batches is 62131 --- greater than max batches
196/1 0.69339 0.68338 +/- 0.00159
Triggers unsatisfied, max unc./thresh. is 18.043133483791827 for absorption in
tally 3
WARNING: The estimated number of batches is 62186 --- greater than max batches
197/1 0.64411 0.68318 +/- 0.00159
Triggers unsatisfied, max unc./thresh. is 18.019303623546417 for absorption in
tally 3
WARNING: The estimated number of batches is 62347 --- greater than max batches
198/1 0.66626 0.68309 +/- 0.00159
Triggers unsatisfied, max unc./thresh. is 17.968092739058083 for absorption in
tally 3
WARNING: The estimated number of batches is 62316 --- greater than max batches
199/1 0.67839 0.68306 +/- 0.00158
Triggers unsatisfied, max unc./thresh. is 17.91968515142146 for absorption in
tally 3
WARNING: The estimated number of batches is 62302 --- greater than max batches
200/1 0.66459 0.68297 +/- 0.00157
Triggers unsatisfied, max unc./thresh. is 17.82970764669685 for absorption in
tally 3
WARNING: The estimated number of batches is 61996 --- greater than max batches
Creating state point statepoint.200.h5...
=======================> TIMING STATISTICS <=======================
Total time for initialization = 2.9309e-01 seconds
Reading cross sections = 2.8108e-01 seconds
Total time in simulation = 1.1321e+01 seconds
Time in transport only = 1.1242e+01 seconds
Time in inactive batches = 1.6721e-01 seconds
Time in active batches = 1.1153e+01 seconds
Time synchronizing fission bank = 2.2958e-02 seconds
Sampling source sites = 1.8701e-02 seconds
SEND/RECV source sites = 3.9403e-03 seconds
Time accumulating tallies = 9.9349e-04 seconds
Total time for finalization = 5.2200e-07 seconds
Total time elapsed = 1.1620e+01 seconds
Calculation Rate (inactive) = 74758.2 particles/second
Calculation Rate (active) = 43708.5 particles/second
============================> RESULTS <============================
k-effective (Collision) = 0.68198 +/- 0.00141
k-effective (Track-length) = 0.68297 +/- 0.00157
k-effective (Absorption) = 0.68161 +/- 0.00145
Combined k-effective = 0.68209 +/- 0.00118
Leakage Fraction = 0.34033 +/- 0.00074
Tally Data Processing¶
[17]:
# We do not know how many batches were needed to satisfy the
# tally trigger(s), so find the statepoint file(s)
statepoints = glob.glob('statepoint.*.h5')
# Load the last statepoint file
sp = openmc.StatePoint(statepoints[-1])
Analyze the mesh fission rate tally
[18]:
# Find the mesh tally with the StatePoint API
tally = sp.get_tally(name='mesh tally')
# Print a little info about the mesh tally to the screen
print(tally)
Tally
ID = 1
Name = mesh tally
Filters = MeshFilter, EnergyFilter
Nuclides = total
Scores = ['fission', 'nu-fission']
Estimator = tracklength
Use the new Tally data retrieval API with pure NumPy
[19]:
# Get the relative error for the thermal fission reaction
# rates in the four corner pins
data = tally.get_values(scores=['fission'],
filters=[openmc.MeshFilter, openmc.EnergyFilter], \
filter_bins=[((1,1),(1,17), (17,1), (17,17)), \
((0., 0.625),)], value='rel_err')
print(data)
[[[0.04508259]]
[[0.0221707 ]]
[[0.10763375]]
[[0.05107401]]]
[20]:
# Get a pandas dataframe for the mesh tally data
df = tally.get_pandas_dataframe(nuclides=False)
# Set the Pandas float display settings
pd.options.display.float_format = '{:.2e}'.format
# Print the first twenty rows in the dataframe
df.head(20)
[20]:
| mesh 1 | energy low [eV] | energy high [eV] | score | mean | std. dev. | |||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| 0 | 1 | 1 | 1 | 0.00e+00 | 6.25e-01 | fission | 2.27e-04 | 1.02e-05 |
| 1 | 1 | 1 | 1 | 0.00e+00 | 6.25e-01 | nu-fission | 5.54e-04 | 2.50e-05 |
| 2 | 1 | 1 | 1 | 6.25e-01 | 2.00e+07 | fission | 7.19e-05 | 1.82e-06 |
| 3 | 1 | 1 | 1 | 6.25e-01 | 2.00e+07 | nu-fission | 1.89e-04 | 4.69e-06 |
| 4 | 2 | 1 | 1 | 0.00e+00 | 6.25e-01 | fission | 2.35e-04 | 9.82e-06 |
| 5 | 2 | 1 | 1 | 0.00e+00 | 6.25e-01 | nu-fission | 5.71e-04 | 2.39e-05 |
| 6 | 2 | 1 | 1 | 6.25e-01 | 2.00e+07 | fission | 6.88e-05 | 1.61e-06 |
| 7 | 2 | 1 | 1 | 6.25e-01 | 2.00e+07 | nu-fission | 1.81e-04 | 4.15e-06 |
| 8 | 3 | 1 | 1 | 0.00e+00 | 6.25e-01 | fission | 2.31e-04 | 1.13e-05 |
| 9 | 3 | 1 | 1 | 0.00e+00 | 6.25e-01 | nu-fission | 5.63e-04 | 2.76e-05 |
| 10 | 3 | 1 | 1 | 6.25e-01 | 2.00e+07 | fission | 6.95e-05 | 1.76e-06 |
| 11 | 3 | 1 | 1 | 6.25e-01 | 2.00e+07 | nu-fission | 1.83e-04 | 4.53e-06 |
| 12 | 4 | 1 | 1 | 0.00e+00 | 6.25e-01 | fission | 2.07e-04 | 9.85e-06 |
| 13 | 4 | 1 | 1 | 0.00e+00 | 6.25e-01 | nu-fission | 5.04e-04 | 2.40e-05 |
| 14 | 4 | 1 | 1 | 6.25e-01 | 2.00e+07 | fission | 6.48e-05 | 1.45e-06 |
| 15 | 4 | 1 | 1 | 6.25e-01 | 2.00e+07 | nu-fission | 1.71e-04 | 3.81e-06 |
| 16 | 5 | 1 | 1 | 0.00e+00 | 6.25e-01 | fission | 2.20e-04 | 1.07e-05 |
| 17 | 5 | 1 | 1 | 0.00e+00 | 6.25e-01 | nu-fission | 5.37e-04 | 2.60e-05 |
| 18 | 5 | 1 | 1 | 6.25e-01 | 2.00e+07 | fission | 6.76e-05 | 1.78e-06 |
| 19 | 5 | 1 | 1 | 6.25e-01 | 2.00e+07 | nu-fission | 1.78e-04 | 4.63e-06 |
[21]:
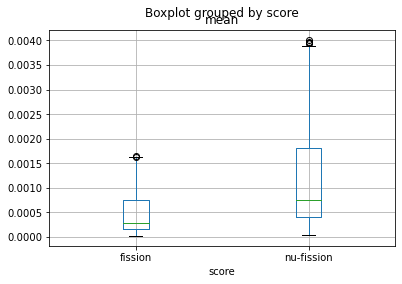
# Create a boxplot to view the distribution of
# fission and nu-fission rates in the pins
bp = df.boxplot(column='mean', by='score')
[22]:
# Extract thermal nu-fission rates from pandas
fiss = df[df['score'] == 'nu-fission']
fiss = fiss[fiss['energy low [eV]'] == 0.0]
# Extract mean and reshape as 2D NumPy arrays
mean = fiss['mean'].values.reshape((17,17))
plt.imshow(mean, interpolation='nearest')
plt.title('fission rate')
plt.xlabel('x')
plt.ylabel('y')
plt.colorbar()
[22]:
<matplotlib.colorbar.Colorbar at 0x7fd070ca32b0>
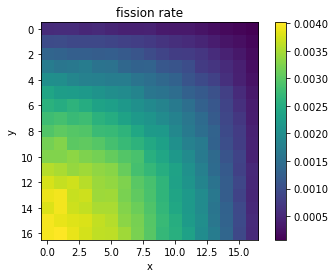
Analyze the cell+nuclides scatter-y2 rate tally
[23]:
# Find the cell Tally with the StatePoint API
tally = sp.get_tally(name='cell tally')
# Print a little info about the cell tally to the screen
print(tally)
Tally
ID = 2
Name = cell tally
Filters = CellFilter
Nuclides = U235 U238
Scores = ['scatter']
Estimator = tracklength
[24]:
# Get a pandas dataframe for the cell tally data
df = tally.get_pandas_dataframe()
# Print the first twenty rows in the dataframe
df.head(20)
[24]:
| cell | nuclide | score | mean | std. dev. | |
|---|---|---|---|---|---|
| 0 | 1 | U235 | scatter | 3.81e-02 | 4.13e-05 |
| 1 | 1 | U238 | scatter | 2.34e+00 | 2.41e-03 |
Use the new Tally data retrieval API with pure NumPy
[25]:
# Get the standard deviations the total scattering rate
data = tally.get_values(scores=['scatter'],
nuclides=['U238', 'U235'], value='std_dev')
print(data)
[[[2.41367509e-03]
[4.12533801e-05]]]
Analyze the distribcell tally
[26]:
# Find the distribcell Tally with the StatePoint API
tally = sp.get_tally(name='distribcell tally')
# Print a little info about the distribcell tally to the screen
print(tally)
Tally
ID = 3
Name = distribcell tally
Filters = DistribcellFilter
Nuclides = total
Scores = ['absorption', 'scatter']
Estimator = tracklength
Use the new Tally data retrieval API with pure NumPy
[27]:
# Get the relative error for the scattering reaction rates in
# the first 10 distribcell instances
data = tally.get_values(scores=['scatter'], filters=[openmc.DistribcellFilter],
filter_bins=[tuple(range(10))], value='rel_err')
print(data)
[[[0.0131914 ]]
[[0.01252949]]
[[0.01241481]]
[[0.01194961]]
[[0.01186091]]
[[0.0127257 ]]
[[0.01358576]]
[[0.0130368 ]]
[[0.014031 ]]
[[0.0141883 ]]]
Print the distribcell tally dataframe
[28]:
# Get a pandas dataframe for the distribcell tally data
df = tally.get_pandas_dataframe(nuclides=False)
# Print the last twenty rows in the dataframe
df.tail(20)
[28]:
| level 1 | level 2 | level 3 | distribcell | score | mean | std. dev. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| univ | cell | lat | univ | cell | |||||||
| id | id | id | x | y | id | id | |||||
| 558 | 3 | 4 | 2 | 7 | 16 | 1 | 3 | 279 | absorption | 7.11e-04 | 1.10e-05 |
| 559 | 3 | 4 | 2 | 7 | 16 | 1 | 3 | 279 | scatter | 8.91e-02 | 6.60e-04 |
| 560 | 3 | 4 | 2 | 8 | 16 | 1 | 3 | 280 | absorption | 6.75e-04 | 1.02e-05 |
| 561 | 3 | 4 | 2 | 8 | 16 | 1 | 3 | 280 | scatter | 8.35e-02 | 6.11e-04 |
| 562 | 3 | 4 | 2 | 9 | 16 | 1 | 3 | 281 | absorption | 6.10e-04 | 1.02e-05 |
| 563 | 3 | 4 | 2 | 9 | 16 | 1 | 3 | 281 | scatter | 7.75e-02 | 6.10e-04 |
| 564 | 3 | 4 | 2 | 10 | 16 | 1 | 3 | 282 | absorption | 5.67e-04 | 9.88e-06 |
| 565 | 3 | 4 | 2 | 10 | 16 | 1 | 3 | 282 | scatter | 7.11e-02 | 5.99e-04 |
| 566 | 3 | 4 | 2 | 11 | 16 | 1 | 3 | 283 | absorption | 5.06e-04 | 9.35e-06 |
| 567 | 3 | 4 | 2 | 11 | 16 | 1 | 3 | 283 | scatter | 6.39e-02 | 5.53e-04 |
| 568 | 3 | 4 | 2 | 12 | 16 | 1 | 3 | 284 | absorption | 4.35e-04 | 8.22e-06 |
| 569 | 3 | 4 | 2 | 12 | 16 | 1 | 3 | 284 | scatter | 5.62e-02 | 5.18e-04 |
| 570 | 3 | 4 | 2 | 13 | 16 | 1 | 3 | 285 | absorption | 3.73e-04 | 7.90e-06 |
| 571 | 3 | 4 | 2 | 13 | 16 | 1 | 3 | 285 | scatter | 4.76e-02 | 4.92e-04 |
| 572 | 3 | 4 | 2 | 14 | 16 | 1 | 3 | 286 | absorption | 2.98e-04 | 7.30e-06 |
| 573 | 3 | 4 | 2 | 14 | 16 | 1 | 3 | 286 | scatter | 3.82e-02 | 4.17e-04 |
| 574 | 3 | 4 | 2 | 15 | 16 | 1 | 3 | 287 | absorption | 2.05e-04 | 5.96e-06 |
| 575 | 3 | 4 | 2 | 15 | 16 | 1 | 3 | 287 | scatter | 2.86e-02 | 3.72e-04 |
| 576 | 3 | 4 | 2 | 16 | 16 | 1 | 3 | 288 | absorption | 1.22e-04 | 4.12e-06 |
| 577 | 3 | 4 | 2 | 16 | 16 | 1 | 3 | 288 | scatter | 1.82e-02 | 2.59e-04 |
[29]:
# Show summary statistics for absorption distribcell tally data
absorption = df[df['score'] == 'absorption']
absorption[['mean', 'std. dev.']].dropna().describe()
# Note that the maximum standard deviation does indeed
# meet the 5e-5 threshold set by the tally trigger
[29]:
| mean | std. dev. | |
|---|---|---|
| count | 2.89e+02 | 2.89e+02 |
| mean | 4.19e-04 | 6.86e-06 |
| std | 2.41e-04 | 2.51e-06 |
| min | 1.68e-05 | 1.07e-06 |
| 25% | 2.06e-04 | 5.09e-06 |
| 50% | 3.98e-04 | 6.90e-06 |
| 75% | 6.17e-04 | 8.44e-06 |
| max | 8.70e-04 | 1.52e-05 |
Perform a statistical test comparing the tally sample distributions for two categories of fuel pins.
[30]:
# Extract tally data from pins in the pins divided along y=-x diagonal
multi_index = ('level 2', 'lat',)
lower = df[df[multi_index + ('x',)] + df[multi_index + ('y',)] < 16]
upper = df[df[multi_index + ('x',)] + df[multi_index + ('y',)] > 16]
lower = lower[lower['score'] == 'absorption']
upper = upper[upper['score'] == 'absorption']
# Perform non-parametric Mann-Whitney U Test to see if the
# absorption rates (may) come from same sampling distribution
u, p = scipy.stats.mannwhitneyu(lower['mean'], upper['mean'])
print('Mann-Whitney Test p-value: {0}'.format(p))
Mann-Whitney Test p-value: 0.47449458604689265
Note that the symmetry implied by the y=-x diagonal ensures that the two sampling distributions are identical. Indeed, as illustrated by the test above, for any reasonable significance level (e.g., \(\alpha\)=0.05) one would not reject the null hypothesis that the two sampling distributions are identical.
Next, perform the same test but with two groupings of pins which are not symmetrically identical to one another.
[31]:
# Extract tally data from pins in the pins divided along y=x diagonal
multi_index = ('level 2', 'lat',)
lower = df[df[multi_index + ('x',)] > df[multi_index + ('y',)]]
upper = df[df[multi_index + ('x',)] < df[multi_index + ('y',)]]
lower = lower[lower['score'] == 'absorption']
upper = upper[upper['score'] == 'absorption']
# Perform non-parametric Mann-Whitney U Test to see if the
# absorption rates (may) come from same sampling distribution
u, p = scipy.stats.mannwhitneyu(lower['mean'], upper['mean'])
print('Mann-Whitney Test p-value: {0}'.format(p))
Mann-Whitney Test p-value: 2.499381683224802e-42
Note that the asymmetry implied by the y=x diagonal ensures that the two sampling distributions are not identical. Indeed, as illustrated by the test above, for any reasonable significance level (e.g., \(\alpha\)=0.05) one would reject the null hypothesis that the two sampling distributions are identical.
[32]:
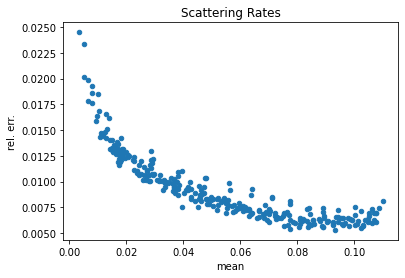
# Extract the scatter tally data from pandas
scatter = df[df['score'] == 'scatter']
scatter['rel. err.'] = scatter['std. dev.'] / scatter['mean']
# Show a scatter plot of the mean vs. the std. dev.
scatter.plot(kind='scatter', x='mean', y='rel. err.', title='Scattering Rates')
<ipython-input-32-c935bd379fee>:4: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
scatter['rel. err.'] = scatter['std. dev.'] / scatter['mean']
[32]:
<AxesSubplot:title={'center':'Scattering Rates'}, xlabel='mean', ylabel='rel. err.'>
[33]:
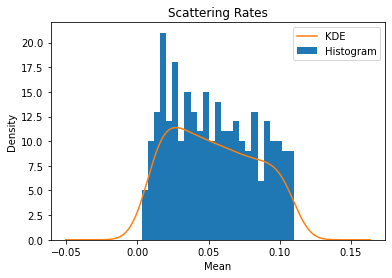
# Plot a histogram and kernel density estimate for the scattering rates
scatter['mean'].plot(kind='hist', bins=25)
scatter['mean'].plot(kind='kde')
plt.title('Scattering Rates')
plt.xlabel('Mean')
plt.legend(['KDE', 'Histogram'])
[33]:
<matplotlib.legend.Legend at 0x7fd070fbd1f0>