Unstructured Mesh: Introduction¶
In this example we’ll look at how to setup and use unstructured mesh tallies in OpenMC. Unstructured meshes are able to provide results over spatial regions of a problem while conforming to a specific geometric features – something that is often difficult to do using the regular and rectilinear meshes in OpenMC.
Here, we’ll apply an unstructured mesh tally to the PWR assembly model from the OpenMC examples.
*NOTE: This notebook will not run successfully if OpenMC has not been built with DAGMC or libMesh support enabled.*
[1]:
from IPython.display import Image
import openmc
import openmc.lib
# ensure one of the two mesh librares is enabled
assert(openmc.lib._dagmc_enabled() or openmc.lib._libmesh_enabled())
We’ll need to download the unstructured mesh file used in this notebook. We’ll be retrieving those using the function and URLs below.
[2]:
%matplotlib inline
from matplotlib import pyplot as plt
plt.rcParams["figure.figsize"] = (30,10)
import urllib.request
pin_mesh_moab_url = 'https://tinyurl.com/u9ce9d7' # MOAB file - 22 MB
pin_mesh_libmesh_url = 'https://tinyurl.com/yysgs3tr' # Exodus file - 9.7 MB
def download(url, filename='dagmc.h5m'):
"""
Helper function for retrieving dagmc models
"""
u = urllib.request.urlopen(url)
if u.status != 200:
raise RuntimeError("Failed to download file.")
# save file as dagmc.h5m
with open(filename, 'wb') as f:
f.write(u.read())
First we’ll import that model from the set of OpenMC examples.
[3]:
model = openmc.examples.pwr_assembly()
We’ll make a couple of adjustments to this 2D model as it won’t play very well with the 3D mesh we’ll be looking at. First, we’ll bound the pincell between +/- 10 cm in the Z dimension.
[4]:
min_z = openmc.ZPlane(z0=-10.0)
max_z = openmc.ZPlane(z0=10.0)
z_region = +min_z & -max_z
cells = model.geometry.get_all_cells()
for cell in cells.values():
cell.region &= z_region
The other adjustment we’ll make is to remove the reflective boundary conditions on the X and Y boundaries. (This is purely to generate a more interesting flux profile.)
[5]:
surfaces = model.geometry.get_all_surfaces()
# modify the boundary condition of the
# planar surfaces bounding the assembly
for surface in surfaces.values():
if isinstance(surface, openmc.Plane):
surface.boundary_type = 'vacuum'
Let’s take a quick look at the model to ensure our changs have been added properly.
[6]:
root_univ = model.geometry.root_universe
# axial image
root_univ.plot(width=(22.0, 22.0),
pixels=(200, 300),
basis='xz',
color_by='material',
seed=0)
[6]:
<matplotlib.image.AxesImage at 0x7f5f451a0860>
[7]:
# radial image
root_univ.plot(width=(22.0, 22.0),
pixels=(400, 400),
basis='xy',
color_by='material',
seed=0)
[7]:
<matplotlib.image.AxesImage at 0x7f5f452f0cf8>
Looks good! Let’s run some particles through the problem.
[8]:
model.run()
%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%
############### %%%%%%%%%%%%%%%%%%%%%%%%
################## %%%%%%%%%%%%%%%%%%%%%%%
################### %%%%%%%%%%%%%%%%%%%%%%%
#################### %%%%%%%%%%%%%%%%%%%%%%
##################### %%%%%%%%%%%%%%%%%%%%%
###################### %%%%%%%%%%%%%%%%%%%%
####################### %%%%%%%%%%%%%%%%%%
####################### %%%%%%%%%%%%%%%%%
###################### %%%%%%%%%%%%%%%%%
#################### %%%%%%%%%%%%%%%%%
################# %%%%%%%%%%%%%%%%%
############### %%%%%%%%%%%%%%%%
############ %%%%%%%%%%%%%%%
######## %%%%%%%%%%%%%%
%%%%%%%%%%%
| The OpenMC Monte Carlo Code
Copyright | 2011-2020 MIT and OpenMC contributors
License | https://docs.openmc.org/en/latest/license.html
Version | 0.12.1-dev
Git SHA1 | e62681221a625ce7eb635df966826379fb4e9453
Date/Time | 2021-01-10 00:51:10
MPI Processes | 1
OpenMP Threads | 2
Reading settings XML file...
Reading cross sections XML file...
Reading materials XML file...
Reading geometry XML file...
Reading U234 from /home/shriwise/opt/openmc/xs/nndc_hdf5/U234.h5
Reading U235 from /home/shriwise/opt/openmc/xs/nndc_hdf5/U235.h5
Reading U238 from /home/shriwise/opt/openmc/xs/nndc_hdf5/U238.h5
Reading O16 from /home/shriwise/opt/openmc/xs/nndc_hdf5/O16.h5
Reading Zr90 from /home/shriwise/opt/openmc/xs/nndc_hdf5/Zr90.h5
Reading Zr91 from /home/shriwise/opt/openmc/xs/nndc_hdf5/Zr91.h5
Reading Zr92 from /home/shriwise/opt/openmc/xs/nndc_hdf5/Zr92.h5
Reading Zr94 from /home/shriwise/opt/openmc/xs/nndc_hdf5/Zr94.h5
Reading Zr96 from /home/shriwise/opt/openmc/xs/nndc_hdf5/Zr96.h5
Reading H1 from /home/shriwise/opt/openmc/xs/nndc_hdf5/H1.h5
Reading B10 from /home/shriwise/opt/openmc/xs/nndc_hdf5/B10.h5
Reading B11 from /home/shriwise/opt/openmc/xs/nndc_hdf5/B11.h5
Reading c_H_in_H2O from /home/shriwise/opt/openmc/xs/nndc_hdf5/c_H_in_H2O.h5
Minimum neutron data temperature: 294.0 K
Maximum neutron data temperature: 294.0 K
Preparing distributed cell instances...
Writing summary.h5 file...
Maximum neutron transport energy: 20000000.0 eV for U235
Initializing source particles...
====================> K EIGENVALUE SIMULATION <====================
Bat./Gen. k Average k
========= ======== ====================
1/1 0.20444
2/1 0.15502
3/1 0.19804
4/1 0.22159
5/1 0.19776
6/1 0.20086
7/1 0.21896 0.20991 +/- 0.00905
8/1 0.23134 0.21706 +/- 0.00885
9/1 0.29029 0.23536 +/- 0.01935
10/1 0.20094 0.22848 +/- 0.01649
Creating state point statepoint.10.h5...
=======================> TIMING STATISTICS <=======================
Total time for initialization = 2.1129e+00 seconds
Reading cross sections = 2.0943e+00 seconds
Total time in simulation = 1.1595e-01 seconds
Time in transport only = 1.1219e-01 seconds
Time in inactive batches = 5.8272e-02 seconds
Time in active batches = 5.7678e-02 seconds
Time synchronizing fission bank = 2.3802e-04 seconds
Sampling source sites = 1.3992e-04 seconds
SEND/RECV source sites = 2.9613e-05 seconds
Time accumulating tallies = 1.4423e-05 seconds
Time writing statepoints = 3.0105e-03 seconds
Total time for finalization = 4.0100e-06 seconds
Total time elapsed = 2.2442e+00 seconds
Calculation Rate (inactive) = 8580.51 particles/second
Calculation Rate (active) = 8668.78 particles/second
============================> RESULTS <============================
k-effective (Collision) = 0.25094 +/- 0.02010
k-effective (Track-length) = 0.22848 +/- 0.01649
k-effective (Absorption) = 0.21556 +/- 0.04156
Combined k-effective = 0.20707 +/- 0.01965
Leakage Fraction = 0.79200 +/- 0.03382
[8]:
PosixPath('/home/shriwise/opt/openmc/openmc/examples/jupyter/statepoint.10.h5')
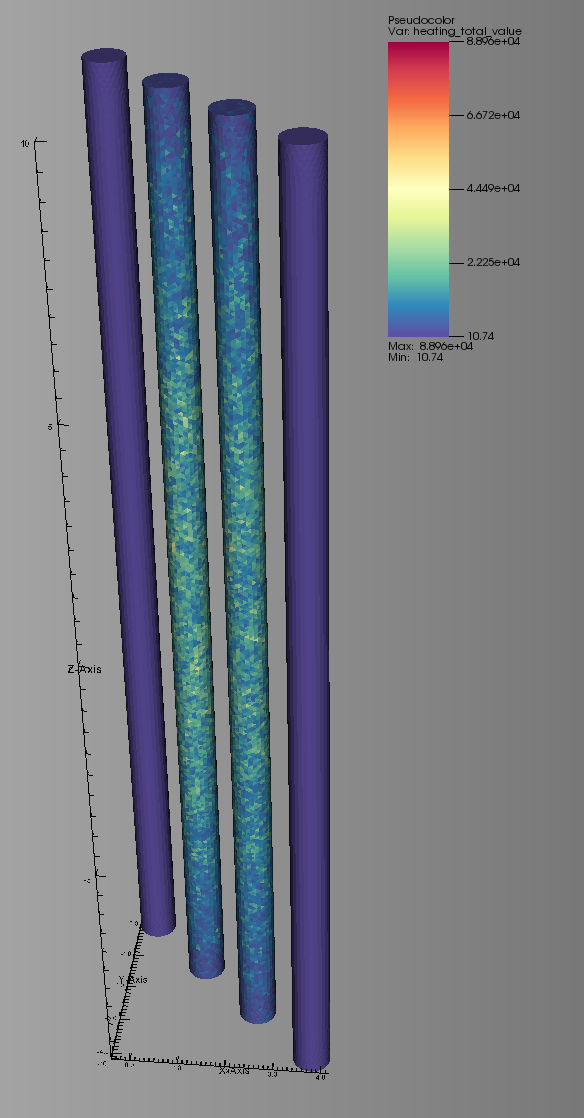
Now it’s time to apply our mesh tally to the problem. We’ll be using the tetrahedral mesh “pins1-4.h5m” shown below:
[9]:
Image("./images/pin_mesh.png", width=600)
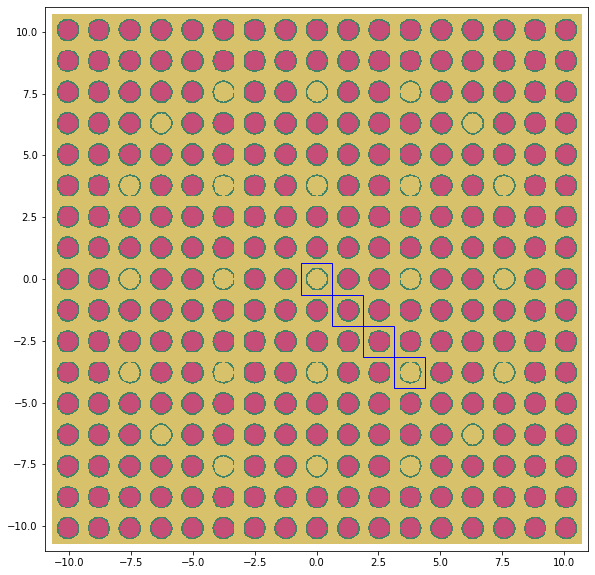
This mesh was generated using Trelis with radii that match the fuel/coolant channels of the PWR model. These four channels correspond to the highlighted channels of the assembly below.
Two of the channels are coolant and the other two are fuel.
[10]:
from matplotlib.patches import Rectangle
from matplotlib import pyplot as plt
pitch = 1.26 # cm
img = root_univ.plot(width=(22.0, 22.0),
pixels=(600, 600),
basis='xy',
color_by='material',
seed=0)
# highlight channels
for i in range(0, 4):
corner = (i * pitch - pitch / 2.0, -i * pitch - pitch / 2.0)
rect = Rectangle(corner,
pitch,
pitch,
edgecolor='blue',
fill=False)
img.axes.add_artist(rect)
Applying an unstructured mesh tally¶
To use this mesh, we’ll create an unstructured mesh instance and apply it to a mesh filter. We do this by specifying a mesh file and mesh library on an UnstructuredMesh object. The specified mesh library will be used to load the mesh file during simulation initialization. OpenMC must be built with support for the specified mesh library enabled.
[11]:
mesh_library = 'moab' # change to 'libmesh' to use libMesh instead
if mesh_library == 'moab':
assert(openmc.lib._dagmc_enabled())
mesh_file = 'pins1-4.h5m'
mesh_url = pin_mesh_moab_url
elif mesh_library == 'libmesh':
assert(openmc.lib._libmesh_enabled())
mesh_file = 'pins1-4.e'
mesh_url = pin_mesh_libmesh_url
# download the file and create the UnstructuredMesh object
download(mesh_url, mesh_file)
umesh = openmc.UnstructuredMesh(mesh_file, library=mesh_library)
Regardless of the library used to represent the mesh, we can apply this mesh object in a MeshFilter.
[12]:
mesh_filter = openmc.MeshFilter(umesh)
We can now apply this filter like any other. For this demonstration we’ll score both the flux and heating in these pins.
[13]:
tally = openmc.Tally()
tally.filters = [mesh_filter]
tally.scores = ['heating', 'flux']
# Only collision estimators are supported for
if umesh.library == 'libmesh':
tally.estimator = 'collision'
model.tallies = [tally]
Now we’ll run this model with the unstructured mesh tally applied. Notice that the simulation takes some time to start due to some additional data structures used by the unstructured mesh tally. Additionally, the particle rate drops dramatically during the active cycles of this simulation.
Unstructured meshes are useful, but they can be computationally expensive!
[14]:
model.settings.particles = 100_000
model.settings.inactive = 20
model.settings.batches = 100
model.run(output=False)
[14]:
PosixPath('/home/shriwise/opt/openmc/openmc/examples/jupyter/statepoint.100.h5')
At the end of the simulation, we see the statepoint file along with a file named “tally_1.100.vtk”. This file contains the results of the unstructured mesh tally with convenient labels for the scores applied. In our case the following scores will be present in the VTK:
- flux_total_value
- flux_total_std_dev
- heating_total_value
- heating_total_std_dev
Where “total” represents the nuclide entry in the tally. If a set of nuclides are specified in the tally, a different score will be added to the VTK for each one in addition to “total”.
Currently, an unstructured VTK file will only be generated for tallies if the unstructured mesh is is the only filter applied to that tally. All results for the unstructured mesh tally are present in the statepoint file regardless of the number of filters applied, however.
These files can be viewed using free tools like Paraview and VisIt to examine the results.
[15]:
!ls *.vtk
tally_1.100.vtk
Heating¶
Here is an image of the heating score as viewed in VisIt. Note that no heating is scored in the water-filled channels as expected.
[17]:
Image("./images/umesh_heating.png", width=600)
Statepoint Data¶
[18]:
with openmc.StatePoint("statepoint.100.h5") as sp:
tally = sp.tallies[1]
umesh = sp.meshes[1]
Enough information for visualization of results on the unstructured mesh is also provided in the statepoint file. Namely, the mesh element volumes and centroids are available.
[19]:
print(umesh.volumes)
print(umesh.centroids)
[1.43381086e-04 1.48043747e-04 1.60408339e-04 ... 7.04197023e-05
7.04197023e-05 7.04197023e-05]
[[ 2.88485691 -2.55429784 9.97768184]
[ 2.87565092 -2.60469781 9.8884092 ]
[ 2.85832254 -2.65291228 9.97768184]
...
[ 1.46082175 -1.15569203 -3.62914358]
[ 1.4443143 -1.1321793 -3.65475081]
[ 1.46884412 -1.15657736 -3.68206543]]
The combination of these values can provide for an appoxmiate visualization of the unstructured mesh without its explicit representation or use of an additional mesh library.
We hope you’ve found this example notebook useful!
[20]:
Image("./images/umesh_w_assembly.png", width=600)